We will use modXNA to build a single strand antisense oligonucleotide (ASO) that complements with an RNA target. The visual representation of the ASO is depicted below:
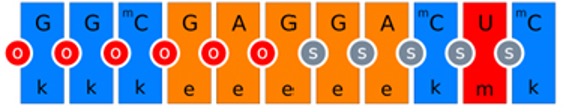
Sketch representation of the ASO. Sugar modifications: Blue is cET (k), red is C2’-O-methyl (m) and orange is C2’-Omethoxyethyl (e). Backbone modifications: red circle between the bases is phosphate linkage and grey circle with an ‘s’ is Phosphorothioate linkage. The mC is 5 methyl cytosine.
ASOs are typically heavily modified and contain a hybrid DNA-RNA-DNA strand. In the example here, the modifications include a 5 methyl cytosine base (M5C) modification, a gapmer (CET) sugar modification, a 2’-O-methoxyethyl (MOE) sugar modification, a 2’-O-methyl (OME) sugar modification, and a phosphorothioate (PS1) backbone modification. To build the ASO, we need the above modifications combined into the following new nucleotides: 5CG, CEG, MEC, MOG, MOA, POG, POA, MSC, OMU, 3EC, detailed in the Table below.
| Residue name | Backbone fragment code | Sugar fragment code | Base fragment code | Details of fragments | Notes |
|---|---|---|---|---|---|
| 5CG | 5PO | CET | RGG | 5′ hydroxyl cap backbone + cET + guanine | 5′-terminal |
| CEG | RPO | CET | RGG | Phosphate backbone + cET + guanine | Central residue |
| MEC | RPO | CET | M5C | Phosphate backbone + cET + 5 methyl cytosine | Central residue |
| MOG | RPO | MOE | RGG | Phosphate backbone + MOE + guanine | Central residue |
| MOA | RPO | MOE | RAA | Phosphate backbone + MOE + adenine | Central residue |
| POG | PS1 | MOE | RGG | Phosphorothioate (S) backbone + MOE + guanine | Central residue |
| POA | PS1 | MOE | RAA | Phosphorothioate (S) backbone + MOE + adenine | Central residue |
| MSC | PS1 | CET | M5C | Phosphorothioate (S) backbone + cET + 5 methyl cytosine | Central residue |
| OMU | PS1 | MOE | RUU | Phosphorothioate (S) backbone + O-methyl + uracil | Central residue |
| 3EC | PS1 | CET | M5C | Phosphorothioate (S) backbone + cET + 5 methyl | 3′-terminal cytosine |
All the new residues follow the same procedure:
- Make a new folder (to keep things organized) with the new residue name (i.e. 5CG)
- In the new folder, make a modXNA input file with the fragment code of the backbone, sugar and base (5CG.in.modxna with the codes: 5PO CET DGG, for the residue 5CG)
- Run modXNA (remember to use the –5cap for the 5CG residue and –3cap flag for the 3EC residue): `
$ modXNA .sh -i 5CG.in. modxna -m 5CG --5cap` - Check that the new residues have been built successfully by opening the tmp.opt.pdb structure.
The resulting structures are shown below:

Before we can use these 10 library files, we need to combine all the different LIB files into a single file. This will make it easier to load just one bigger LIB file into LEaP instead of loading 10 independent LIB files. To do this, create the text file (create-ASO-lib.tleap)
loadoff 3EC/3EC.lib
saveoff 3EC ASO.lib
loadoff 5CG/5CG.lib
saveoff 5CG ASO.lib
loadoff CEG/CEG.lib
saveoff CEG ASO.lib
loadoff MEC/MEC.lib
saveoff MEC ASO.lib
loadoff MOA/MOA.lib
saveoff MOA ASO.lib
loadoff MOG/MOG.lib
saveoff MOG ASO.lib
loadoff MSC/MSC.lib
saveoff MSC ASO.lib
loadoff OMU/OMU.lib
saveoff OMU ASO.lib
loadoff POA/POA.lib
saveoff POA ASO.lib
loadoff POG/POG.lib
saveoff POG ASO.lib
quitRun the file with;
$ tleap -s -f create-ASO-lib.tleap
This will load all the LIB files we created with modXNA and save the information into the same ‘ASO.lib’ file. If you inspect the ASO.lib file generated by tleap, you will see that all the new residues are in one single file.
Now that we have all the required nucleotides, we need to build the ASO. There are several methodologies that we can use to proceed. The simplest one is generating a single strand with the correct sequence of nucleotide analogs using the ‘sequence’ command available in LEaP, using the following script (Aso-single-strand.tleap)
###########################
source leaprc.DNA.OL24
source leaprc.RNA.OL3
source leaprc.water.opc
##########################
loadoff ASO.lib
loadamberparams /home/user/modXNA/dat/frcmod.modxna
##########################
ASO=sequence{5CG CEG MEC MOG MOA MOG POG POA MSC OMU 3EC}
solvateoct ASO OPCBOX 9.0
addionsrand ASO Na+ 0
addionsrand ASO Cl− 0
saveamberparm ASO ASO−ss.parm7 ASO−ss.crd
quitLEaP will create a linearized structure with the correct sequence of residues. It will have some structural overlaps, however, and if we run a quick structural minimization using AMBER/sander (using the 9-step protocol from Amber-MDPrep), we obtain a structure with no clashes:
