Example on how to use modXNA script and collection of fragments to build a non-canonical nucleotide fragment.
- Pre-requisites
- Confirm that AmberTools is installed in your system
- Confirm that you have the latest version of CPPTRAJ
- Important considerations
- modXNA does not build 5′- or 3′- terminal base pairs.
- modXNA is designed to provide an initial working molecule (nucleotide) that can be further refined with new parameters if needed.
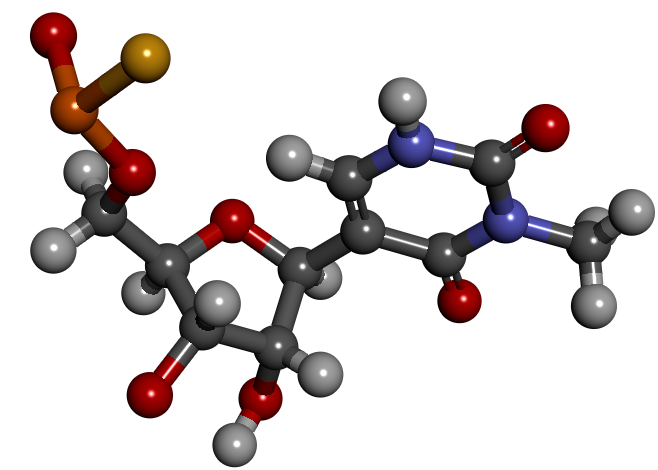
In this example we are going to build a simple nucleotide that has a phosphothioate backbone and a modified base (3-methylpseudouridine).
- Download the latest version of modXNA
- Untar the file:
tar xvfz modXNA-<latest-version>.tar.gz
- modXNA requires an input file (text file) that indicates what fragments to join. The text file has the form:
- <backbone ID> <sugar ID> <base ID>
where each ID for each fragment can be obtained from the CATALOG of fragments.
- <backbone ID> <sugar ID> <base ID>
- For this example we need:
- the phosphothioate backbone – fragment ID: PS1
- the sugar, we will use ribose – fragment ID: RC3
- the base, we will use 3-methylpseudouridine – fragment ID: M3Y
- With the fragment ID’s, we write the input text file (with name in.modxna):
PS1 RC3 M3Y
- We now run modXNA:
modxna.sh -i in.modxna
The script will run and perform several tasks:
– Generate a random name for the new residue.
– Grab the fragments from the dat/ path.
– Delete the declared capping groups.
– Use CPPTRAJ to graft together the fragments using the declared HEAD/TAIL atom mask for each fragment.
– Use CPPTRAJ to adjust the charge.
– Run a short geometry minimization using SANDER.
– Generate an AMBER compatible library file that can be loaded in LEaP.
Below is an example script for loading modXNA library and frcmod files in tleap:
## Amber OL3 force field is used to parameterize
## canonical nucleotides
source leaprc.RNA.OL3
source leaprc.water.opc
## Load modxna frcmod & library files - the random
## three-letter code ("SWR") generated from modxna MUST ## match the residue name in the structure PDB
loadamberparams modxna/dat/frcmod.modxna
loadoff lib/SWR.lib
## Load and solvate the system
rna = loadpdb start.pdb
addions rna Na+ 0
addions rna Cl- 0
solvateoct rna OPCBOX 10.0
## Add additional ions for appropriate environment, in ## this case our volume is 182273.67 A^3 and we would
## like a 200 mM NaCl concentration
addions rna Na+ 22
addions rna Cl- 22
saveamberparm rna rna.topo rna.coords